The Challenge
Functional medicine practitioners spend nearly 40% of their time analyzing complex patient data from multiple sources (blood tests, genetic reports, diet logs). Manually correlating this data to provide precision nutrition advice is error-prone and time-consuming.
The Goal: Build an AI-powered assistant that can ingest diverse medical documents, "understand" the patient's unique biological context, and provide evidence-based clinical recommendations in real-time.
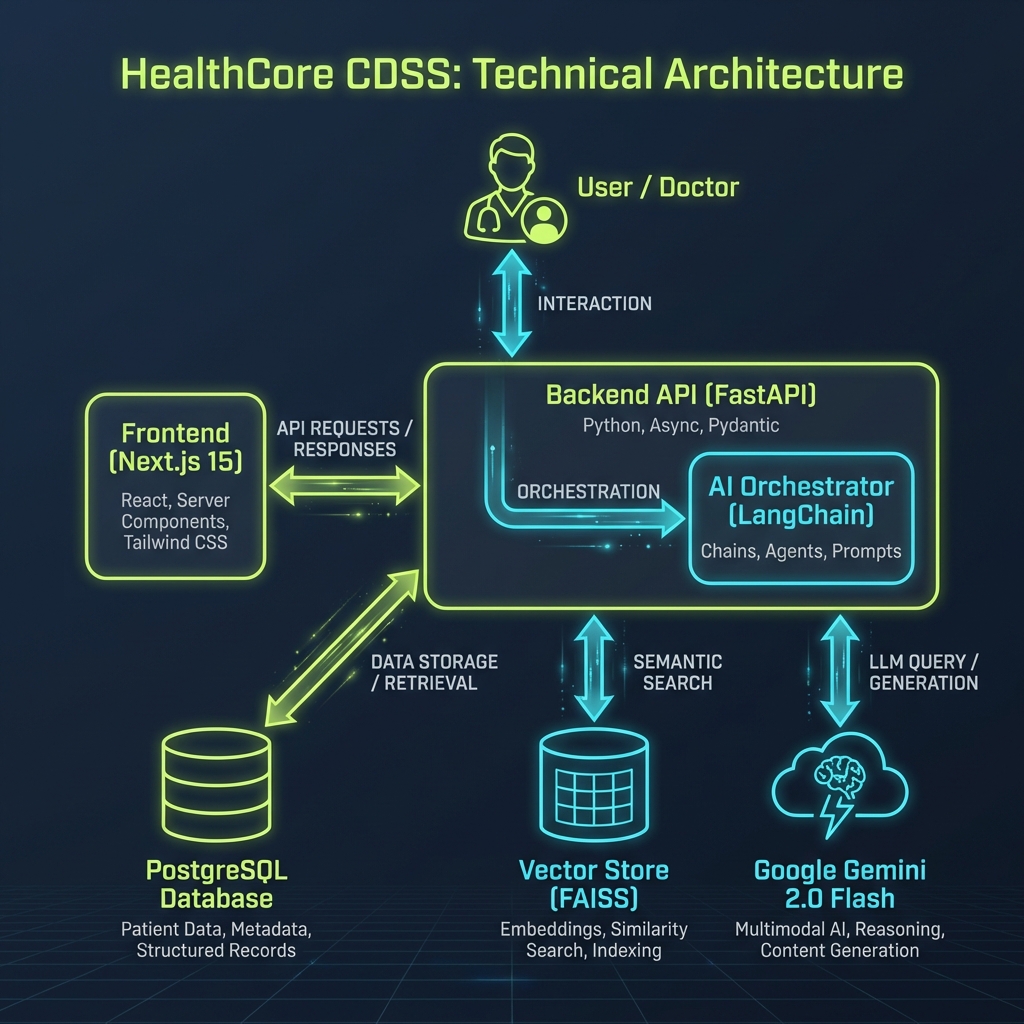
System Architecture
HealthCore is built on a modern microservices-ready architecture designed for scalability and data privacy.
1. Backend & AI Orchestration
The core logic resides in a FastAPI backend. I chose Python for its robust ecosystem in AI/ML. The system uses LangChain to orchestrate interactions between the LLM and our data sources.
2. RAG (Retrieval-Augmented Generation) Pipeline
Instead of fine-tuning a model (which is costly and rigid), I implemented a RAG architecture:
- Ingestion: Medical PDFs constitute unstructured data. I built a custom parser to extract text while preserving clinical context.
- Embedding: Text chunks are converted into vector embeddings.
- Retrieval: When a user asks "What should this patient eat?", the system retrieves relevant patient history + medical protocols from the vector store (FAISS).
- Generation: The retrieved context + the query are fed into Google Gemini 3.0 to generate a grounded, accurate response.
💡 Why RAG instead of Fine-Tuning?
Medical data changes rapidly. Fine-tuning a model is expensive and results in static knowledge. RAG (Retrieval-Augmented Generation) allows the system to be dynamic: simply updating the vector database instantly gives the AI access to the latest research and patient records without retraining. This ensures real-time accuracy and zero hallucination on critical data.
Key Technical Challenges
Prompt Engineering for Clinical Safety
One of the biggest risks in Health AI is hallucination. To mitigate this, I engineered a "System Prompt" that enforces a strict persona: "You are a functional medicine expert. Only answer based on the provided context. If unsure, state that data is missing." This drastically reduced incorrect advice.
Handling Multilingual Data
The system needed to support Turkish character sets and medical terminology seamlessly. I addressed this by ensuring UTF-8 compliance across the entire ETL pipeline and selecting an embedding model capable of cross-lingual understanding.
Tech Stack Breakdown
| Component | Technology | Role |
|---|---|---|
| frontend | Next.js (React) | Responsive Clinical UI |
| Backend API | FastAPI (Python) | High-performance Async Endpoints |
| AI Orchestration | LangChain | Managing LLM Context & Prompts |
| LLM Model | Google Gemini 3.0 | Medical Reasoning & Generation |
| Vector DB | FAISS / Supabase | Semantic Search Storage |
| DevOps | Docker | Containerization |
Future Roadmap
The project is currently in production beta. Upcoming features include:
- Agentic Workflow: Moving from a passive QA bot to an autonomous agent that can proactively suggest follow-up tests.
- Multi-modal Input: Analyzing food photos directly using Vision models.